Inhibit inhibition: PCR inhibition and how to prevent it

Polymerase chain reaction (PCR) is established as a rapid, sensitive, and specific molecular biological tool for the analysis of nucleic acids. Today, PCR is utilized for genetic analysis of a wide range of samples, diagnosis of infectious or hereditary disorders, in forensic science, and for food and water quality analysis, among other purposes. PCR variants include the reverse transcription PCR (RT-PCR), using RNA as template, which is initially transcribed into DNA by a reverse transcriptase, and real-time or quantitative PCR (qPCR), which uses fluorescent probes for the detection of the PCR product providing quantitative information. The PCR is a chemical enzymatic reaction in which several components play an essential role in the DNA amplification process. Efficient in vitro DNA polymerization needs high DNA polymerase activity as well as favorable nucleic acid interactions (target denaturation and primer annealing), implying that both biochemical and biophysical processes are involved. A compound affecting any of the critical reagents or subreactions in the polymerization process acts as an inhibitor. The inhibitors’ impact can range from partial inhibition and underestimation of the target nucleic acid amount to complete amplification failure. There are numerous reasons for a deficient PCR. If you are convinced that your chemicals are in good condition, you have added the PCR reagents in the appropriate concentration (primers, dNTP’s, MgCl2, and Taq polymerase) as well as the template DNA you want to amplify, but still obtain a blank on your PCR, it is most likely that there are inhibiting components in the reaction.
Inhibitor origin
PCR inhibitors are a heterogeneous class of substances and the most common reason for amplification failure when DNA yield is sufficient. They can derive either from the original sample, from sample preparation prior to PCR, or from both.
1. Inhibitors coming from sample collection.
PCR inhibitors can be found in a variety of biological materials analyzed in clinical, food, and environmental testing. Clinical samples, such as blood, serum or plasma, urine, and stool, contain several substances that can affect PCR amplification. For example, native immunoglobulin (IgG) present in blood is one of the most potent PCR inhibitors since it has exceptional affinity for single-stranded DNA. You can partially remove this inhibitory effect by incubating the sample with a nonspecific DNA to neutralize the IgG. Regarding food samples, you should be aware of certain components that can compromise your PCR efficiency. In milk, the contained enzyme plasmin degrades the Taq polymerase and high calcium concentration might inhibit the PCR. If you work with plants, you might have experienced that they carry many inhibitory substances. Polysaccharides, like dextran sulphate, xylan, dextransulphate, and gutti gum affect the PCR efficiency. Moreover, phenols present in berries and tomatoes seem to especially inhibit RT-PCR assays. On the other hand, dead biomass and soil may contain humic and fulminic acids, which inhibit PCR even at low concentrations. In sewage sludge and wastewater, we can find fats, proteins, polyphenols, and heavy metals, polysaccharides, metal ions, and RNases – all of them are common PCR inhibitors of environmental samples. For completeness, other inhibitors include constituents of bacterial cells, non-target DNA and contaminants, and laboratory items such as pollen, glove powder, laboratory plasticware, and cellulose.
2. Inhibitors introduced during extraction process.
We know that several substances may be necessary for efficient cell lysis or for the preparation of pure nucleic acids, but they may cause PCR inhibition at certain concentrations. Those include different salts (sodium or potassium chloride), ionic detergents, non-ionic detergents or organic molecules (EDTA, ethanol, isopropyl alcohol or phenol). Ionic detergents (e.g., sodium deoxycholate, sarkosyl and SDS) are highly inhibitory for the PCR, whereas non-ionic detergents (e.g., Nonidet P-40, Tween 20, Triton X-100 and N-octyl glucoside) cause PCR inhibition only at relatively high concentrations. EDTA is found in several elution buffers of purification kits for preservation of DNA, but at certain concentrations, it may deplete magnesium ions and thus inhibit your DNA polymerase activity. Phenol likes to denature proteins and can inhibit enzymes in your downstream applications; thus, you should ensure your final product does not contain phenol impurities. While ethanol is an important part of the protocol's washing steps, it is also a strong PCR inhibitor. It is critical to let all the ethanol evaporate before eluting the DNA. Some protocols recommend lithium chloride for RNA desalting because it is more soluble in ethanol than sodium acetate. However, be cautious because it can inhibit protein synthesis and DNA polymerase. Additives of the PCR mixture, such as dithiothreitol, dimethyl sulphoxide or mercaptoethanol, may also be inhibitory at certain concentrations. Generally, the material of swabs or the composition of transport media may also influence the sensitivity of the PCR.
Mechanisms of inhibition
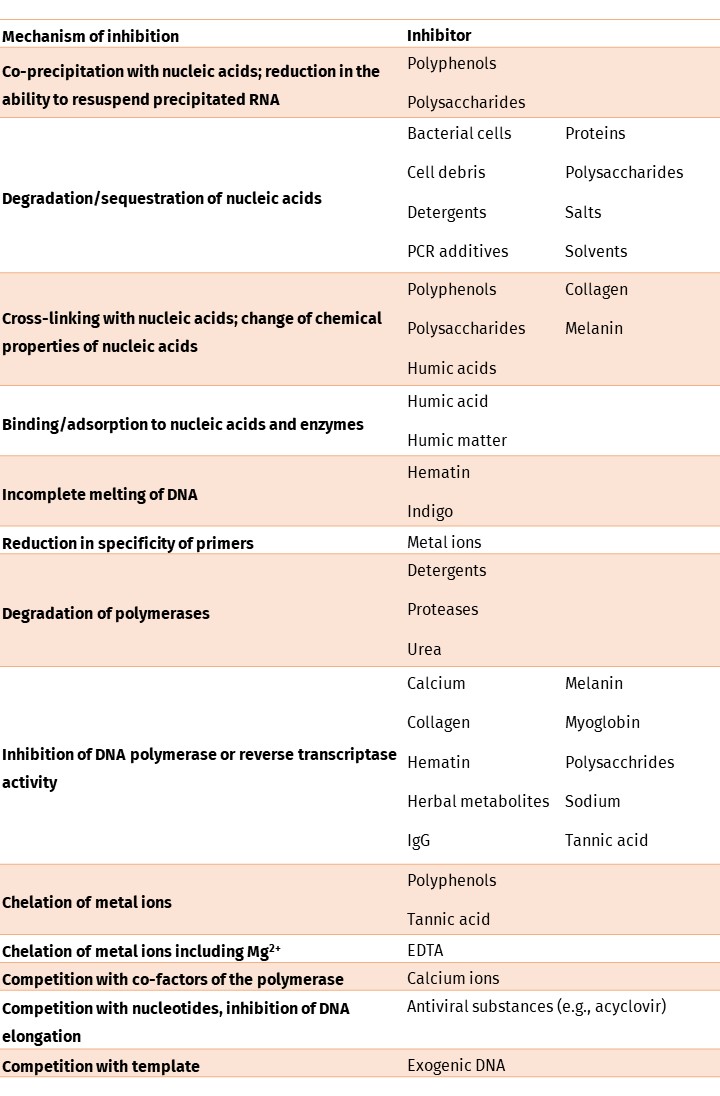
The efficacy of the PCR or qPCR performance may be affected by different inhibitors interfering at different stages of the process. Despite for many components it is not well understood how they exert its inhibition, hereinbelow, we refer to the most critical steps where they can operate (extended list in Table 1).
Some inhibitors escape removal during the purification procedure by binding directly to single- or double-stranded nucleic acids (Figure 1A). For example, nucleases may modify or degrade template RNA or DNA, and phenols may cross-link RNA under oxidizing conditions and thus hamper RNA isolation. In addition, the existence of polysaccharides may reduce the capacity to resuspend precipitated RNA. Besides, DNA polymerases have cofactor requirements that can be the target of inhibition (Figure 1B). High concentrations of calcium may lead to a competitive binding by the DNA polymerase instead of magnesium, and complexing agents, like tannic acids, deplete magnesium. In both cases, magnesium is no longer available as cofactor for the polymerase and its activity is decreased.
Furthermore, DNA polymerase can also be degraded, blocked, or altered (Figure 1B). For example, proteases or detergents present in the reaction, urea, and phenol can degrade this enzyme. Calcium, collagen, hematin, and tannic acid may inhibit polymerase activity. Melanin forms a reversible complex with the DNA polymerase and polysaccharides may disturb the enzymatic process by mimicking the structure of nucleic acids. Humic acids interact with the template DNA and the polymerase thus preventing the enzymatic reaction even at low concentrations.
Additionally, annealing of the primers to the DNA template may be hampered by certain PCR inhibitors (Figure 1C). As this effect is caused by a competitive binding of the inhibitor to the template, you can overcome this problem with an appropriate primer design leading to higher melting points.
Other substances inhibit reverse transcription by directly binding to the reverse transcriptase, which results in its denaturalization, or binding to the DNA nascent chain, which leads to a premature transcription termination (Figure 1D). For example, melanin inhibits reverse transcription by direct interaction with the enzyme.
Finally, in real-time PCR assays, interference with the fluorescent probes or increased background fluorescence represent additional inhibition mechanisms (Figure 1E).
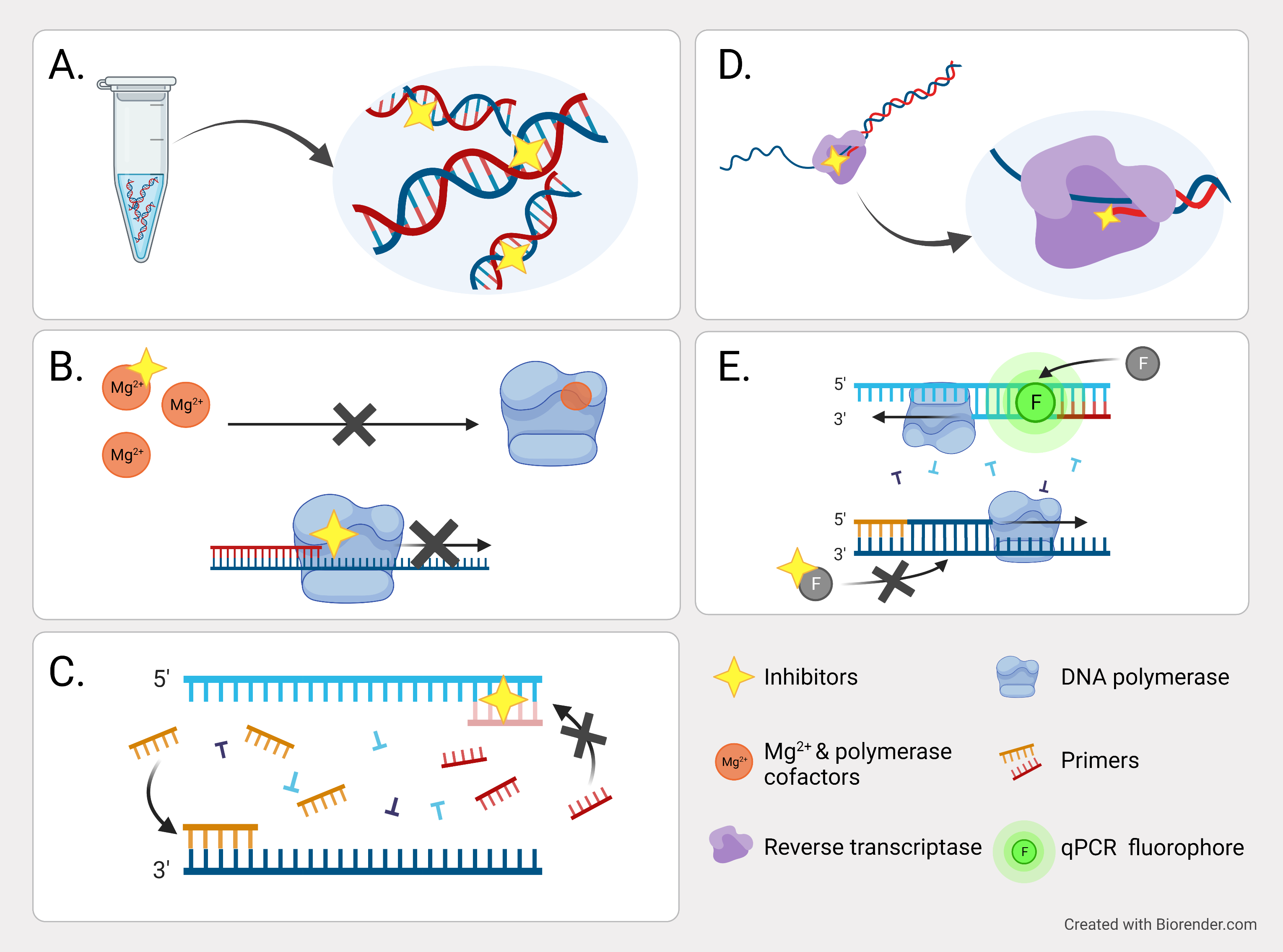
Figure 1. Representation of the attack points of PCR inhibitors during sample preparation and PCR procedure. A. Inhibitors can react to double or single- stranded nucleic acids and co-precipitate during the purification process. B. Some substances reduce Mg2+ and other polymerase cofactors availability, which interfere with DNA activity. C. Inhibitors interfere with primers annealing. D. Some agents bind to the reverse transcriptase or bind to the DNA chain and terminates it. E. Inhibitors can prevent the fluorophores binding to the nascent DNA.
Table 1. Common inhibitors derived from different sample types and mechanisms of inhibition.
Methods for removal of PCR inhibitors
In general, you can mitigate the effects of the inhibitors by using an appropriate sample processing and nucleic acid extraction method, by the choice of a more robust DNA polymerase, or employing specific PCR additives.
1. General methods.
In our previous article we described several techniques extensively used to remove a wide range of PCR inhibitors from DNA and RNA samples: phenol-chloroform, ethanol precipitation, silica columns, cation exchange resins, magnetic beads, and single-step purification (EchoLUTION ). Another general method is the dilution of the sample or the extracted nucleic acids, which will automatically result in a dilution of the PCR inhibitors to a level that does not affect the reaction. It is a simple strategy, but the effectiveness may depend on the nature of the inhibitor. The downside is that since you are diluting your target, you also decrease sensitivity.
2. DNA polymerase selection.
Thermostable DNA polymerase is a key component in the amplification reaction, and any factor interfering with the enzymatic activity will affect the amplification capacity. The DNA polymerase can be degraded, denatured, or have its enzymatic activity reduced by a wide variety of compounds present in biological samples. We recommend choosing an appropriate DNA polymerase and, if applicable, an appropriate reverse transcription system according to your sample type. For example, the commonly used Taq polymerase is considerably less resistant to blood (being completely inhibited by 0.004 % (vol/vol)) compared with DNA polymerases isolated from Thermus thermophilus (rTth polymerase) and Thermus flavus (Tfl polymerase), which exhibit non reduced efficiency in the presence of 20 % (vol/vol) blood. Several Taq polymerases have been generated by site-directed mutagenesis resulting in greater resistance to inhibitors in blood and soil as well as an increased tolerance to high concentrations of DNA-binding dyes. For example, mutants of Taq polymerase with a higher affinity for primer-template (Kd 1.1 nM for mutant versus 102 nM for wild-type Taq). By fusing a DNA polymerase to a ssDNA binding domain, the inhibitor tolerance is also improved for human blood, lactoferrin, and heparin. We are not going to cover all the existing alternatives, but it is important to know that several DNA polymerases are commercially available. These polymerases exhibit very different properties regarding resistance to various components in biological samples and performance in the presence of these components.
3. Amplification facilitators.
During the development of PCR methodology, the basic master mixture containing DNA polymerase, primers, nucleotides, and a reaction buffer (Tris-HCl, KCl, and MgCl2) has been supplemented with numerous compounds to enhance the amplification efficiency. Such compounds are called amplification facilitators. They can influence amplification at various stages and under different conditions by increasing or decreasing the thermal stability of the DNA template, influencing the error rate of the DNA polymerase, affecting system specificity, and relieving amplification inhibition caused by complex biological samples.
- Proteins. The two proteins that are most frequently used to facilitate amplification are BSA and the single-stranded DNA-binding protein gp32, which is a protein encoded by gene 32 of bacteriophage T4. BSA can help you to overcome PCR inhibition caused by blood- or heme-containing substances, phenolics, humic, and tanic acid among others, by binding these components. The degradation of DNA polymerase by proteinases present in fecal samples can cause amplification inhibition. BSA and gp32 have been proposed to alleviate this inhibition effect by acting as a target for proteinases.
- Organic solvents. Dimethyl sulfoxide (DMSO) and formamide are two organic solvents frequently used as PCR facilitators. Both solvents might influence the thermal stability of the primers and the thermal activity profile of the DNA polymerase, thereby increasing the specificity of amplification.
- Nonionic detergents. The main nonionic detergents used as PCR facilitators are Tween-20 and Triton X, which stimulate the activity of Taq DNA polymerase and reduces false terminations of the enzyme.
- Biologically compatible solutes. Betaine and glycerol are the most common facilitators, which are used by organisms and cellular systems to maintain biological activity under extreme conditions. For that reason, you can use glycerol in the storage buffer of thermostable enzymes. It facilitates amplification by enhancing the hydrophobic interactions between protein domains and lowering the strand separation temperature of DNA. The addition of both betaine and glycerol to amplification reaction mixes will help you to reduce the formation of secondary structures caused by GC-rich regions.
- Polymers. PEG and dextran can facilitate amplification in similar ways as organic solvents. Also, PEG was reported to relieve the inhibition caused by feces and the plant polysaccharide dextran sulfate.
Conclusion
PCR inhibitors are a heterogeneous class of substances that act at different steps of the analysis. They are present in a large variety of sample types and may lead to decreased PCR sensitivity or even false-negative PCR results. Several strategies have been developed to remove PCR inhibitors during sample preparation. But since it cannot be guaranteed that the preparations are free of PCR inhibitors, all reactions should be analyzed for the presence of inhibitory effects. Standardized controls are therefore recommended to achieve reliable PCR results and to compare efficiencies of different PCR protocols. Stay tuned for our next blog article.
To avoid PCR inhibitors during sample processing we advise using an appropriate nucleic acid extraction method. BioEcho offers a variety of nucleic acid extraction kits for a wide variety of samples. Our innovative EchoLUTION technology ensures nucleic acids of high yield and free of inhibitors for reliable downstream applications.

