The perfect sequence: no-fail DNA extraction methods
Extracting the perfect sequence
Next-generation sequencing (NGS) is used to determine the nucleotide sequence of specified DNA sections or complete genomes with extremely high throughput, scalability and speed. NGS has revolutionized the biological sciences, allowing labs to obtain genetic information from biological samples more rapidly, accurately, and cheaper than ever before. As such, NGS is a crucial technique for unravelling a growing number of scientific and diagnostic challenges for all sorts of organisms, tissues, and samples. Although library preparation methods for NGS have become highly robust, the quality and quantity of the DNA input still has a major impact on the reliability and usability of the generated data. As such, it is essential to use high molecular weight (HMW) DNA that is of high quality and concentration to get the most out of your sequencing. This requires the most efficient method of DNA extraction, correct handling and storage of your sample, and preservation of the sample's integrity. DNA degradation, and biological and chemical contaminants represent a few examples of many factors which may confound the optimal outcome due to improper storage and isolation. To ensure that storage and extraction conditions are effective, it is critical that the methods used are targeted towards the specific sample type as each sample will have its own requirements. In this blog we will cover the fundamentals of choosing the best methods for preparing your samples for NGS, making sure to consider the impact of sample type on storage and DNA extraction.
Why does your sample type matter?
Each biological sample type comes with its own unique challenges associated with sample degradation and DNA extraction. For instance, metabolically active tissues such as the liver or kidney have high enzymatic activity and will degrade DNA quickly if not frozen or stabilized. In contrast, blood samples are relatively stable and can be left for up to a week at room temperature without impacting DNA quality.
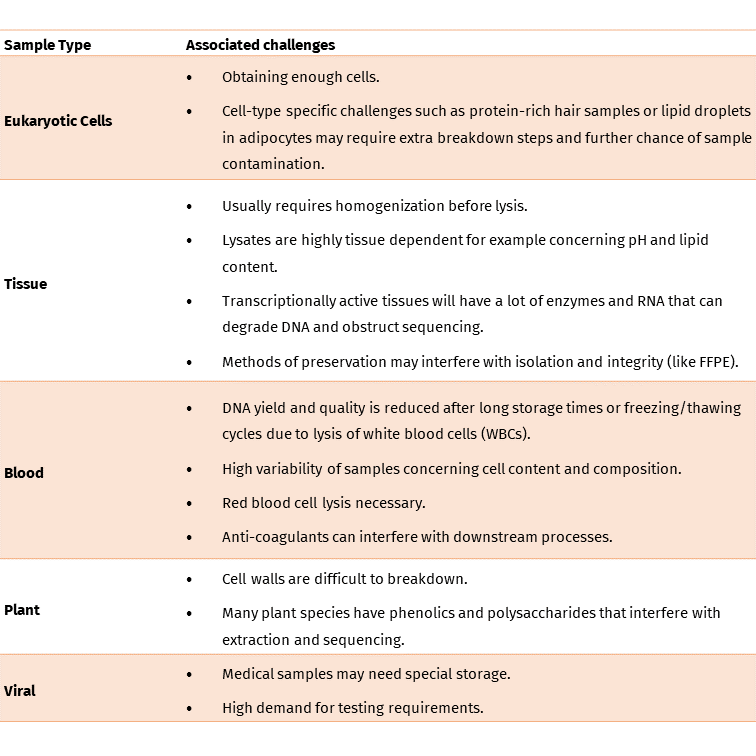
The following table shows potential difficulties related to each sample type:
As such, your sample type will likely determine what approach you use for sample storage and also DNA extraction.
Sample storage
Sequencing adheres to the “garbage in, garbage out” rule; therefore, it is wise to take care when preparing your samples. Since enzymes that can break down DNA are released as soon as samples are harvested, it is often important to start the DNA extraction process quickly to ensure that the DNA you are isolating is intact. Due to this, the highest DNA yield and quality is generally achieved by purifying genomic DNA from freshly harvested tissues and cells. However, in many cases, samples cannot be processed immediately after harvesting and so it is important to know how best to preserve your biological sample for when you are ready to extract.
Eukaryotic cells
To prevent ice crystal formation and disruption of cell structure, you could consider using a medium containing a cryopreservation agent such as DMSO - however, these can interfere with downstream processes like PCR and sequencing.
Optimum practice for storing cells is to centrifuge harvested cell cultures, remove the buffer and then store the cells at –20°C or –80°C.
Tissue
Needs to be frozen immediately or stabilized:
If freezing, consider homogenizing your sample beforehand for easier DNA extraction later. Homogenization can be achieved through several techniques such as grinding, bead beating, or mixing. Your choice will depend on the sample toughness and access to equipment such as liquid nitrogen or tissue lysing machines.
If stabilizing, consider using ethanol or a stabilizing agent such as PurifyLater Tissue Stabilizer. This allows for easier handling and manipulation of the tissue sample which may be needed if it is to be cut up into smaller fragments.
While FFPE is a good method for fixing tissue, it can result in significant fracturing and denaturing of DNA and thus, it is not suitable for producing HMW DNA for sequencing.
Blood
Whole blood samples can be stored in the fridge for up to one week or if long term storage is needed, at –20 °C or –80 °C.
If lacking storage space, the WBCs and platelets can be separated from the whole blood sample through centrifugation and stored as a buffy coat. However, due to the high cell count, this may impact DNA extraction later.
Generally, heparin should be avoided, as it may interfere with downstream processes. However, some kits like EchoLUTION Blood Micro Kit can still process samples stabilized with heparin.
Plant
Plant tissue can be stored at 4 °C if DNA extraction is completed in less than 24 hours, but storage at -80 °C is necessary for any duration greater than that.
For long-term storage, it may be best to use liquid nitrogen to grind the tissue into a powder to be stored at -80 °C.
If freezing is not an option, the sample should be dried immediately using lyophilizers, silica gel, or food dehydrators. For long-term storage, dried samples can be kept in the dark, at room temperature, in dry or air-sealed conditions.
Virus
Swabs and stool samples containing viral DNA should be immediately frozen at -80 °C.
Choosing the right DNA extraction method
With the right storage method sorted, you are ready to extract DNA from your samples. The extraction process involves the breakdown of the extracellular matrix and cell membrane using mechanical force, enzymes, solvents, or surfactants. DNA can then be isolated from the resulting mixture. It is essential that the DNA extraction results in highly pure DNA as other molecules, such as RNA, proteins and chemical contaminants, may interfere with the downstream fragmentation, PCR and sequencing processes. Furthermore, the genomic DNA should be of good integrity and checked for double stranded DNA content. The best DNA extraction method will ideally be tailored to your sample type and compatible with your storage method.
There are several different major DNA extraction methods, including organic, non-organic methods, and newer methods available as commercial kits:
Organic extraction: This method is the traditional way to extract DNA from cells and tissues but is time consuming and involves the use of hazardous chemicals. Cells are disrupted by the organic solvents, phenol and chloroform, and then centrifuged to form a biphasic solution. DNA from the top layer is then precipitated by adding high concentrations of salt, such as sodium acetate, and ethanol or isopropanol.
Non-organic extraction: Often used with CTAB (Cetyl Trimethyl Ammonium Bromide) to help break down cell walls and separate polysaccharides when extracting DNA in plants, these techniques achieve DNA purification through protein precipitation at high salt concentration. Similar to organic extraction, these protocols can also be lengthy and repetitive with low throughput, involving many steps that require careful handling and exposure to toxic chemicals.
Commercial kit methods: Many commercial kits involve either a spin column or magnetic beads. Principally, DNA binds to either the column or the beads in both techniques. Afterwards, there are several washing steps to remove impurities and salts. Although both methods are quite rapid and simple, there are still some drawbacks such as the large number of washing steps which increases loss in yield, and the fact that contaminants are often still carried over, especially in plant samples, resulting in the need for further purification steps.
Learn more in our blog post on DNA extraction techniques.
Extraction IV: A New Hope for Sequencing
Understandably, this may leave you less than clear about which DNA extraction method to use! Although some have clear advantages for your sample type, most have drawbacks on lengthy protocols, hazardous reagents and potential yield and purity issues that would hinder downstream sequencing.
However, there is hope,
Firstly, if you have been using the techniques previously described, it is possible to further purify your DNA post-extraction. Clean-up kits, such as EchoCLEAN remove inhibitors and impurities from DNA samples in a single step to improve the results of your downstream applications. However, a better solution would be a better DNA extraction method to begin with.Fortunately, another highly-effective technique has been recently developed to extract DNA in a single-step purification process - the EchoLUTION workflow. This no-fail method is similar to the aforementioned spin column technique in that a column is used to separate DNA from other cellular components.
However, in contrast to this method, EchoLUTION yields pure DNA as it is the DNA that flows through while everything else stays firmly stuck to the column. Additionally, because no chaotropic salts or organic solvents are used in this procedure, the eluted DNA is free of any possible impurities, making it ideal for the requirements of downstream sequencing.
Thanks to the unique single-step purification, this method also reduces time and costs while increasing the purity and yield of DNA.And the great news is that this method is effective no matter your sample or storage technique - with kits that are tailored to each sample type.
For example, extracting DNA from plants is usually tricky and requires multiple steps due to the tough cell wall and insoluble compounds. However, just one centrifugation step can elute high quality, pure DNA. Additionally, kits specifically for extracting DNA from blood samples are effective with fresh, frozen and stabilized samples, including those stabilized with heparin which is usually very difficult to separate from the DNA sample. To get the most of your sequencing, it is best to have HMW DNA that is of excellent quality and concentration. In order to do this, the integrity of your sample must be preserved through proper handling and storage, and DNA must be extracted using the most effective technique. But whichever DNA extraction method you choose, it is important not to underestimate the impact of tailoring to your sample type to extract the perfect sequence.
At BioEcho, we are pioneers of nucleic acid extraction technology. Learn more about our innovative extraction methods. Looking for a customized extraction service? Contact us for more information.