Clean up your act: DNA and RNA purity
RNA, DNA

In our previous articles, we highlighted how crucial is to obtain highly pure nucleic acid material to improve the results of downstream applications, such as PCR, cloning, genotyping, next-generation sequencing (NGS) and RNA sequencing (RNA-seq) or transcriptomics.
Impurities in the nucleic acid preparation could lead to inhibition or to inaccurate results. The purity of your DNA or RNA will depend on the initial material and the extraction method, though most of them still carry some impurities on the final output. In many cases, additional cleanup is necessary. Here, we talk about nucleic acid purity and how to get a clean nucleic acid outcome.
Get rid of impurities in DNA and RNA samples
Based on the type of starting material, the extraction protocol varies. However, the basis of a standard extraction procedure consists of two main steps. First, a lysate from the initial material (animal or plant cells, tissues, blood) is created. Second, the lysate is cleared to separate the genetic material from unwanted materials, which are cell debris (proteins, lipids, and other molecules) and substances used during the procedure (detergents, reagents, enzymes). Purification of nucleic acids with standard methods takes several steps. It starts with the nucleic acid binding to a matrix or with phase separation, followed by washing steps, and the final elution of the DNA/RNA. Most conventional methods encounter difficulties with full removal of certain impurities obtained through the isolation procedure such as salts, detergents, enzymes, and organic solvents. Furthermore, an additional DNA refining process is necessary for PCR amplification products, and from different enzymatic reactions using restriction enzymes, ligases, kinases, etc. RNA needs cleanup after purification methods such as phenol/chloroform-based extraction and after enzymatic reactions such as in vitro transcription, DNase I treatment, capping, and labeling.
Conventional methods to clean up your DNA and RNA samples
After your nucleic acid extraction, we recommend to carefully measure purity through A230/A260 and A280/A260 ratios and to observe the spectra absorbance. These data will give you an interpretation of the contaminants you might have in your throughput and based on that you can decide the most suitable method to clean up your sample. Most of the methods for nucleic acid cleanup are based on the same principle as extraction methods. Hereinbelow, we review some of those methods:
1. Phenol-chloroform approach is the traditional procedure to remove proteins from a DNA sample. This method consists in mixing the DNA sample with phenol and chloroform, commonly followed by ethanol precipitation. The hydrophilic DNA separates into the aqueous phase, while the proteins, as denature in the presence of organic solvents, remain in the organic phase, or lie at the phase interface.
After phenol-chloroform RNA extraction, samples might carry over phenol as it is partially soluble and stays in the aqueous phase (which can contain up to 7 % phenol). Phenol likes to denature proteins and can inhibit enzymes in your downstream applications. Pure chloroform works nicely to extract phenol from aqueous solutions. You just need to add 1 volume chloroform, vortex and take off the upper layer. The step can be repeated as many times as necessary.

2. Ethanol precipitation is a standard and effective method for desalting and concentrating DNA or RNA. A concentration of 70 % ethanol is added to your sample, together with monovalent cations (acetate salt or sodium at 0.1 to 0.5 M). Most salts and organic solvents are soluble at 70 % ethanol, while nucleic acid molecules aggregates and precipitates. Nucleic acid precipitates can then be easily separated by centrifugation (Figure 1B). A commonly used salt is sodium acetate for routine DNA and RNA precipitation. However, you can apply sodium chloride (NaCl) for samples containing SDS, since NaCl maintains SDS soluble in 70 % ethanol and does not precipitate with the DNA. Some protocols recommend lithium chloride for RNA desalting because it is more soluble in ethanol than sodium acetate. But beware that can inhibit protein synthesis and DNA polymerase. Ammonium acetate is an alternative to remove dNTPs.

3. Anion exchange results in high purity nucleic acids for downstream applications. Column chromatography has evolved to provide a rapid and effective alternative to more laborious methods for preparing high-quality DNA, such as CsCl-gradient centrifugation. The technique is based on positively charged diethylaminoethyl (DEAE) resins, which interact with the negatively charges of the nucleic acid backbone. The salt concentration and pH conditions of the buffers used in each step control binding, subsequent wash to remove contaminants such as proteins and cellular debris, and selective nucleic acid elution from the resin (Figure 1C). There are different column types available on the market with different binding capacities (indicated in µg) for different classes of nucleic acids. For example, the capacity for double-stranded plasmid DNA is 100 µg, and for RNA it is twice as high. Large nucleic acids such as genomic DNA are bound to a slightly lower capacity than plasmid DNA. This relationship between the binding capacity of the resin and the size of the nucleic acids being prepared must be considered when calculating expected yields.

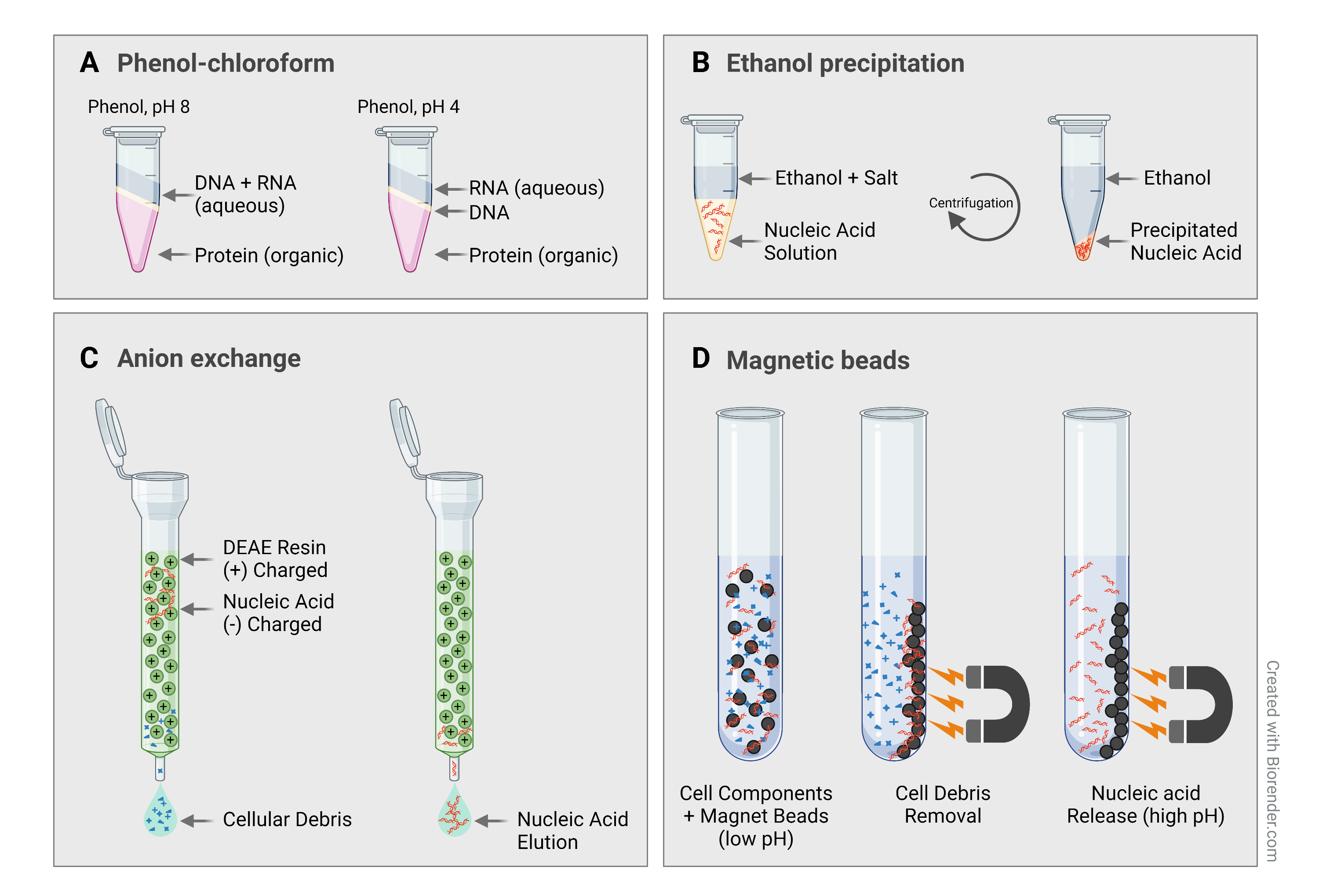
Figure 1. Methods to cleanup your DNA and RNA samples


Inconvenience of common methods for nucleic acid cleanup
Phenol-chloroform extraction and ethanol precipitation approaches are time-consuming in comparison to other available methods, as ethanol precipitation is an overnight step. Alternatively, ethanol precipitation at 80 °C for 30 minutes could reduce procedure time. Moreover, you risk phenol-chloroform and ethanol carry over into the final throughput, which could inhibit downstream applications. Plus, phenol-chloroform is a hazardous component and generates harmful residues that need to be proceeded. It should be always handled in a hood as it is a toxic agent.
Resins from the anion exchange method and magnetic beads can be expensive. Additionally, the aspiration step when using magnetic beads can be tricky and both, beads, and sample, can be lost during the process.
Available cleanup kits based on the silica membrane technology claim no buffer retention and no carryover of contaminants. During DNA purification with column systems, in which a plastic ring fixes the membrane in the column, ethanol residues frequently enter the eluate. This inhibits enzymatic steps, and DNA can float up when agarose gels are loaded. Chaotropic salt carryover could, however, be an issue, as these are added to the sample to allow nucleic acids binding to the silica membrane. Moreover, silica-based kits follow a bind/wash/elute workflow (Figure 2) that needs time to proceed the samples and potentially loses some amount of the precious nucleic acid fragments.
We already talked about how we can increase purity, but this can be at expenses of integrity. If the sample, however, is not clean enough, most likely you will compromise your downstream application. We recommend evaluating what your needs are according to the subsequent applications.
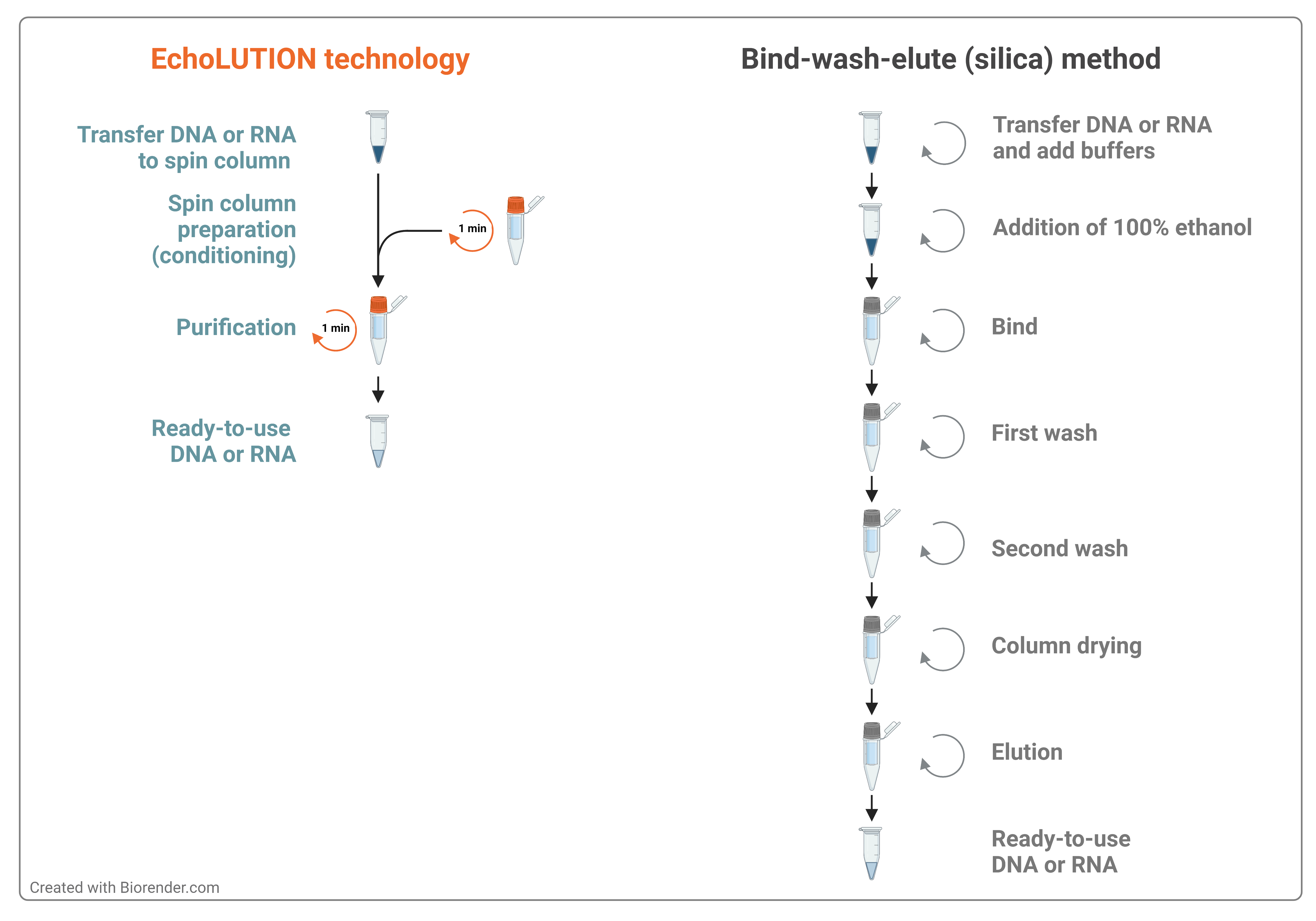
Convenient and efficient cleanup method
BioEcho Life Sciences has developed a series of unique EchoCLEAN cleanup kits that allows to obtain nucleic acids of superior purity in a single centrifugation step. EchoCLEAN kits not only lead to a drastically reduced processing time, but also deliver a high recovery of the target nucleic acid. We offer solutions for the purification of different kinds of impurities: organic solvents (phenol, Trizol, chloroform, ethanol), salts (chaotrophs, SDS, Sodium), dyes (Indigo, gel loading), and any others (primers, dNTPs, precipitates). Check out the EchoCLEAN portfolio and select a kit based on your needs.

