Measuring DNA - And Why It Might Be Wrong
RNA, DNA

If you’ve ever worked in a molecular biology lab there’s a high chance that measuring DNA was a common part of your workflow. But have you ever thought about how accurate your quantification methodology is?
While numerous DNA quantification approaches exist (Table 1) there is no universally accepted gold standard. However, as we discover in this blog, that doesn’t mean that every DNA quantification technique is right for your downstream application. Let’s find out why measuring DNA is an important step in your quality control process – and discuss why it might be wrong.
Why is it important to measure DNA?
Following nucleic acid extraction, it is always useful to know what the DNA yield is. Not only does this demonstrate that your extraction was successful, but it is sometimes necessary to determine the volume needed for downstream applications, such as PCR. And let’s not forget that most quantification methods also give an indication of purity – an invaluable piece of information for applications such as next generation sequencing (NGS).
Inaccurate
quantification of DNA can not only hinder optimization of your
molecular biology workflow but, in some cases, ruin your entire
experiment. For example, in NGS accurate DNA quantification is needed to
ensure the right size sequencing library is input into the sequencer.
If the sequencing library is underestimated, for example, the sequence
coverage will be reduced resulting in a lower degree of confidence.
Ultimately, inaccurately measuring DNA can lead to sub-optimal results!
Spectrophotometric analysis
One of the most commonly used methods for measuring DNA is by spectrophotometric analysis. A spectrophotometer not only determines the average concentrations of nucleic acids in a sample but also their purity.
The
technology exploits the fact that both DNA and RNA absorb ultraviolet
light at 260 nm. Spectrophotometers measure the intensity of a
monochromatic light that passes through a sample and reaches a detector.
To measure nucleic acid concentration, it calculates the 260 nm
absorbance. Based on the Lambert Beer Law, in simple terms the amount of
light absorbed is proportional to the concentration of DNA/RNA in the
sample.
But does spectrophotometric analysis measure up?
To
explore this, genomic DNA was extracted from rat brain samples using
two different kits: (1) A commercially available silica column-based
extraction and (2) BioEcho’s EchoLUTION Tissue DNA Micro Kit. Total DNA yield was then quantified by both spectrophotometry at 260 nm (OD260 value) and agarose gel for analysis.
The
spectrophotometric results suggested that higher DNA yields were
achieved using the silica column kit. However, gel analysis tells a
different story. The gel images show that short, contaminating DNA/RNA
fragments in the silica column samples are responsible for the high and
misleading spectrophotometric readings (Fig. 1). In fact, highest genomic DNA yields are achieved with the EchoLUTION kit.
This experiment illustrates a fundamental limitation of DNA quantitation by spectrophotometry: it lacks specificity. Spectrophotometers measures anything absorbing at 260 nm—whether that be DNA, RNA, protein, free nucleotides or excess salts. Therefore, if you rely exclusively on spectrophotometry, you could be dramatically overestimating your DNA which may have detrimental consequences on your downstream applications.
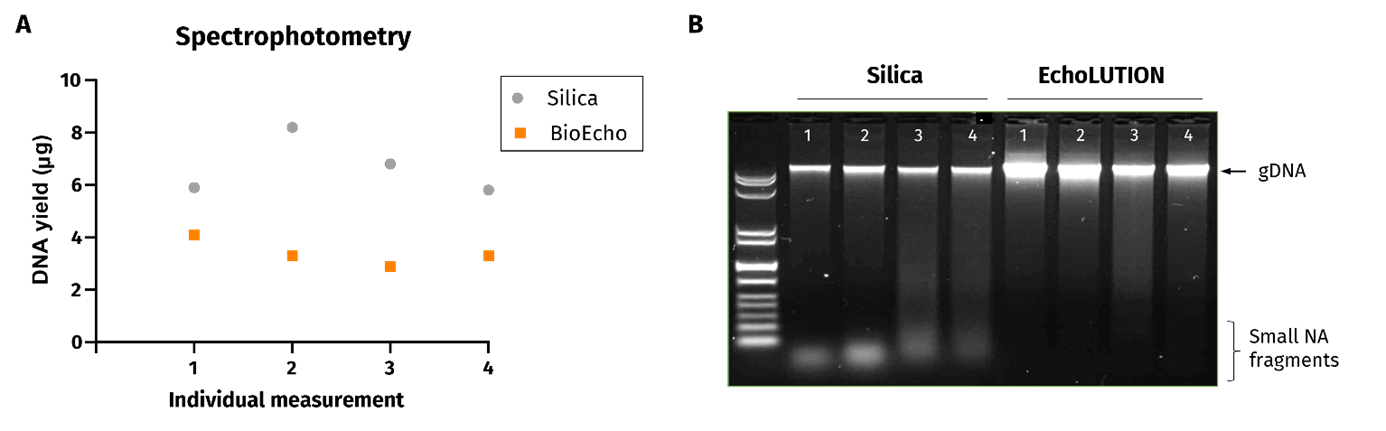
Figure 1. Comparison of DNA quantification techniques analyzed by spectrophotometry and electrophoresis gels. A. DNA extracted from mouse brain samples using (1) a commercially available silica column-based extraction kit and (2) EchoLUTION Tissue DNA Micro Kit from BioEcho.
DNA yields were quantified by spectrophotometry. B. Gel analysis of
aforementioned samples. DNA amounts of around 100 ng per well were
loaded and visualized with GelRed®.
Measuring DNA… What’s the alternative?
Your optimal DNA quantification technique will depend on application. For instance, quantitative PCR (qPCR) and new droplet digital PCR technologies have proven popular for quantitating sequencing libraries. This is because they are sensitive methods that use PCR primers designed to specifically target library adaptor sequences. Consequently, PCR technologies deliver accurate results on the pool of nucleic acids important to sequencing (i.e., adaptor-ligated sequences).
However,
freshly extracted DNA does not contain adaptor sequences. Fluorometers,
such as Qubit, offer a more sensitive approach compared to
spectrophotometers for measuring total DNA yield – even at yields as low
as 10 pg/µL.
Fluorometry uses fluorescent dyes that only emit
when bound to target molecules. They are estimated to be orders of
magnitude more sensitive than UV absorbance and unlike spectroscopy, can
selectively target either DNA, RNA or proteins. However, they may also
deliver misleading results if the DNA extraction method yields samples
containing contaminating DNA (e.g., short DNA fragments in genomic DNA
extractions).
Measuring DNA accurately
Fluorometric methods offer significant improvements on spectrophotometric methods for measuring DNA. However, if you really want to ensure accurate DNA quantification, a robust extraction kit that yields contaminant-free, high-quality DNA is essential.
Due to its unspecific nucleic
acid binding method, silica-based extraction kits purify any nucleic
acid - including small unwanted DNA and RNA molecules. New advances –
such as EchoLUTION technology – are improving nucleic acid extractions,
yielding targeted, high molecular weight DNA in high yields and free
from inhibitors and contaminating RNA/DNA.
Discover more about how EchoLUTION technology can give you high-quality DNA – and save you time
At BioEcho, we’re pioneers of nucleic acid extraction technology. Learn more about our innovative extraction methods. Looking for a customized extraction service? Contact us for more information.

